In Brief:
- Biochemists are finding new applications for the flexibility of ribonucleic acid (RNA).
- New Jersey’s Jonathan Ouellet is researching how to use specific RNA sequences called aptamers to target and kill cancer cells.
- A segment of RNA sequences called a riboswitch interacts with target molecules to regulate and modify gene expression.
Glass panels complemented warm beige walls, making Dr. Jonathan Ouellet’s biochemistry lab at Monmouth University in West Long Branch, New Jersey, more modern and sleeker than any the college freshman had seen. It was as if the work of the scientist were under a spotlight. The new student’s attention skimmed over rows of beakers and flasks of clear liquids and fixed on a mysterious machine on the corner of the counter. He had no idea that the different combinations of ribonucleic acid, or RNA, right in front of his eyes could one day help to cure cancer.
Focusing on RNA
RNA is an intermediate molecule found between double-stranded DNA and the production of protein. It is essential in the genetic instructions for life and development in living organisms. Because it is a single-stranded molecule, RNA can fold into countless three-dimensional, characteristic shapes. Taking its flexibility and reactivity into account, Dr. Jonathan Ouellet is focusing on RNA in his preliminary research to develop a potential cure for cancer.
Locating Cancer Cells
Cancer cells leave behind a unique chemical trace called target molecules. Dr. Ouellet is making specific RNA sequences called aptamers that bind to the chemical trace to locate cancer cells. Aptamers are perfect for the job because their different individual shapes allow for high precision in binding with target molecules.
Not all aptamers occur naturally, so Dr. Ouellet’s team of researchers needs to manually “design” each aptamer sequence. Although there are virtually endless combinations, the team is narrowing down this previously tedious process using the SELEX artificial evolution method. This method analyzes billions of aptamer segments and tests their abilities to bind to the target molecule. When a certain aptamer sequence proves to be effective, researchers can fine-tune the shape for more accuracy in detecting the cancer cells.
Modifying Gene Expression
Dr. Ouellet’s team is enlisting the help of Mother Nature to destroy cancer cells. Gene expression, or the process of making proteins from RNA, is a natural process in living organisms. By manipulating this process, scientists can get their desired end product using a cell’s existing machinery. To do this, scientists use the riboswitch, another segment of RNA sequences that contains a unique aptamer.
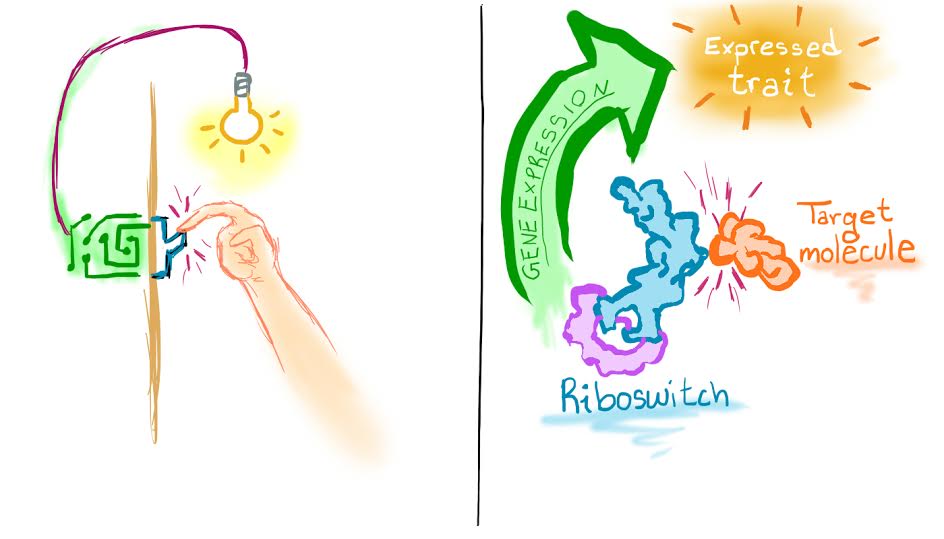
Because the pathways of gene expression are different in cancerous cells than in normal cells, researchers hope to “switch on” gene expression to create a toxic protein that can kill the cancer cells. In healthy cells, the riboswitch remains turned off and unaffected by the cell’s activity.
While Dr. Ouellet’s team is in the early stages of the project, the researchers are getting closer to their ambitious vision every day. As for the college freshman, the immense potential of the study has sparked his enthusiasm for volunteer work at the lab and opened his eyes to the wonders of biochemistry. He has already learned to pay close attention because he almost missed the importance and versatility of the RNA hiding in plain sight in the beakers of Dr. Ouellet’s lab.
Works Cited
- Aquino-Jarquin, Guillermo, and Julia D. Toscano-Garibay. “RNA Aptamer Evolution: Two Decades of SELEction.” IJMS International Journal of Molecular Sciences 12.12 (2011): 9155-171. Web.
- Ellington, Andrew D., and Jack W. Szostak. “In Vitro Selection of RNA Molecules That Bind Specific Ligands.” Nature 346.6287 (1990): 818-22. Web.
- Losman, Julie-Aurore, and William G. Kaelin. “What a Difference a Hydroxyl Makes: Mutant IDH, (R)-2-hydroxyglutarate, and Cancer.” Genes & Development 27.8 (2013): 836-52. Web.
- Soukup, Garrett A., and Ronald R. Breaker. “Nucleic Acid Molecular Switches.” Trends in Biotechnology 17.12 (1999): 469-76. Web.
- Zimmermann, Bob, Ivana Bilusic, Christina Lorenz, and Renée Schroeder. “Genomic SELEX: A Discovery Tool for Genomic Aptamers.” Methods 52.2 (2010): 125-32. Web.
Katelyn McCreedy: Editor-in-Chief
Vivian Qiang: Creative Team Coordinator
Aparna Ragupathi: Copy Editor
Sreya Das: Graphic Designer
Image Credits:
Feature Image: [“Protein-4eyb” by CLS Research Office, (Edited). License: CC BY-SA 2.0]
[“Rabid Mechanical RNA” by Pascal, (Edited). Public Domain]
Story Image: Graphic by Staff Illustrator, Sreya Das
This article was written by Yihan Wu. As always, before leaving a response to this article please view our Rules of Conduct. Thanks! -cYw Editorial Staff
