There are always new viruses emerging, and, like SARS-CoV-2, which causes COVID-19, they are constantly evolving into different variants or strains. Researchers are racing to find vaccines and therapies that can improve outcomes for patients worldwide, using methods ranging from traditional lab work to computational biology (bioinformatics), and even artificial intelligence (AI). To understand vaccine development, we first need to understand how our immune system fights germs.
“The scientific world is constantly on the lookout for potential new pandemics so when there is a new virus, we would be able to quickly predict and measure the immune response,” said Dr. Alessandro Sette, a professor at the La Jolla Institute for Immunology in San Diego, California, and Director of the Center for Cancer Immunotherapy and Center for Vaccine Innovation. Being able to predict and measure how the body’s immune system will respond to viruses is essential to developing effective vaccines. The immune system recognizes, remembers, and destroys disease-causing organisms, called pathogens, and can provide long-lasting protection from future attacks. Pathogens are made up of antigens, which activate the immune response.
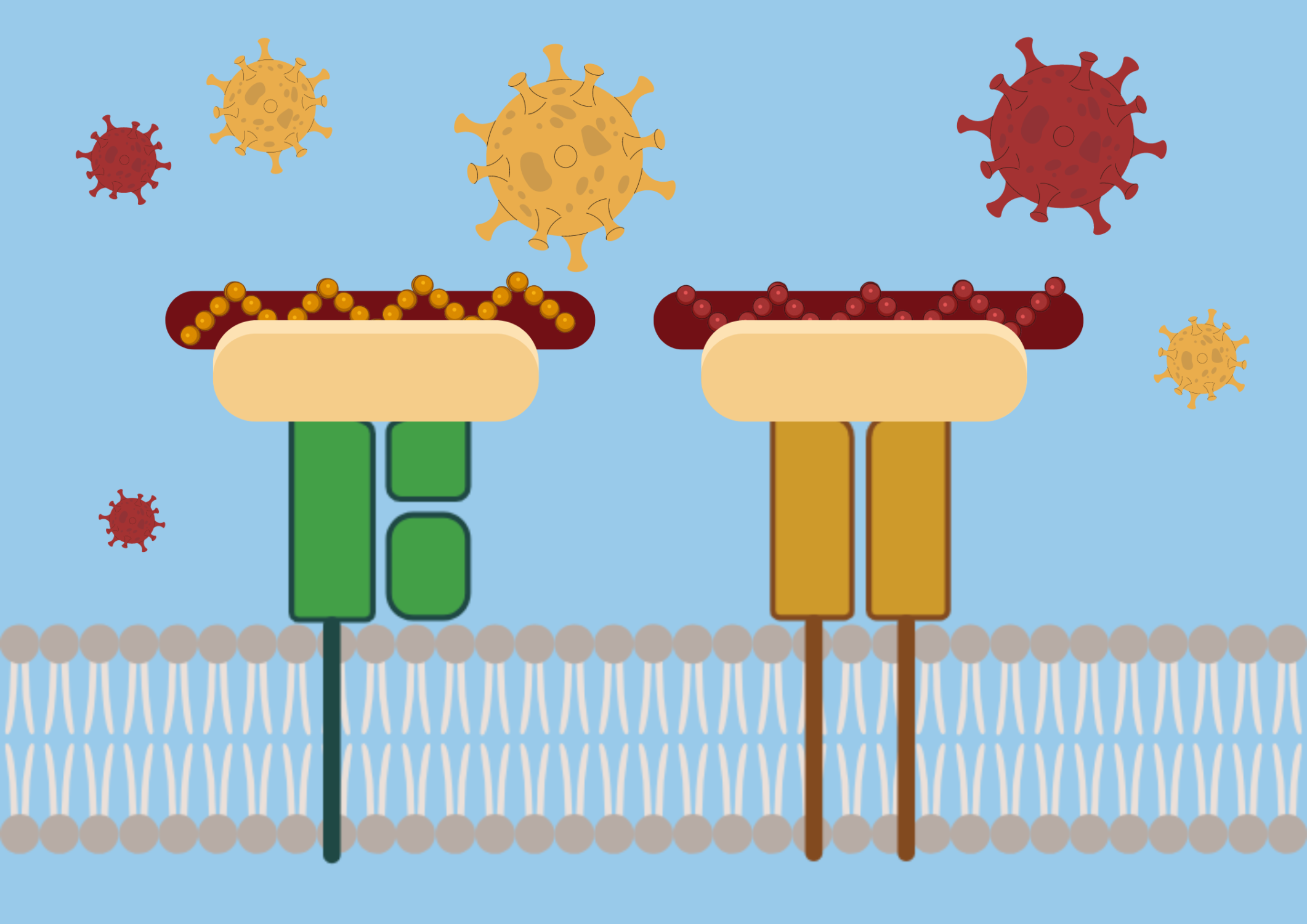
The body’s immune response is mediated by B cells and T cells. They do not recognize pathogens as a whole but instead recognize epitopes, which are unique markers on the antigens. If you’ve ever seen pictures of the SARS-CoV-2 virus, the spikes on the virus’s surface are the antigens that allow researchers to develop COVID-19 vaccines. These are critical for the immune system’s ability to identify and respond to foreign invaders such as viruses and bacteria.
Predicting B-cell vs. T-cell epitopes
The difference between B cells and T cells makes it necessary to have multiple methods for predicting their epitopes. B cells produce antibodies that usually bind to cell-surface epitopes that are folded in a three-dimensional structure. This method used for B-cell epitope prediction is called discontinuous 3D structure-based epitope prediction.
“Antibodies recognize things on the outside and often recognize three-dimensional structures that are made out of discontinuous epitopes,” said Dr. Sette. “These are epitopes that are made from parts of a protein that are not necessarily like ducks in a row.”

T cells are entirely different: They recognize chopped-up fragments of proteins bound to human leukocyte antigen (HLA) molecules. Also known as major histocompatibility complex (MHC), these are specialized molecules on a cell’s surface for detection, holding important epitopes, for T cells. T-cell epitope prediction, therefore, is not limited to far-apart 3D structures like B cells are. Instead of discontinuous epitopes, T cells recognize linear epitopes. This method is called linear sequence-based epitope prediction.
“If you could see the structure of an HLA molecule with a peptide bound to it, it looks like a hot dog bun with a sausage in the middle,” Dr. Sette explained. “That is the fragment where the peptide is stretched out.”
Currently, most vaccines and therapeutics target B cells because antibodies are easier to measure than epitope fragments in T cells, although both are important for vaccine design.
Bioinformatics in advancing epitope prediction
In the past, vaccines were developed by using whole inactivated pathogens (such as in polio), an approach that was not always successful, or by predicting epitopes using traditional lab techniques, which are laborious and time-consuming. Recent advances in computational biology and bioinformatics have significantly improved the ability to predict epitopes for B-cell and T-cell activation in a time-sensitive manner.
Dr. Sette is part of a team that develops and oversees the national Immune Epitope Database (IEDB), a free, widely-used bioinformatics resource database for storing epitope structures. It has two purposes: to function as a catalog for epitopes and as a collection of epitope prediction tools for immunology research around the world. The IEDB uses many methods to predict epitopes and is always being updated. Generally, it analyzes patterns in already-known epitope structures to predict the epitope for an unknown one for B-cell or T-cell activation. One key area in these advances lies in AI. Machine learning (ML) algorithms, trained on large datasets of known epitopes and their interactions, can improve the accuracy of predictions. Dr. Sette plans to use ML approaches to improve data curation and algorithm prediction.
“We will be relying on predictions more than data that is already available because if it is a new virus, we’ll have to rely on more innovative approaches,” Dr Sette said. He believes that if another pandemic arrives, epitope prediction will give researchers an upper hand in fighting against its spread.
- Researchers are using AI and bioinformatics to develop vaccines by predicting how the immune system fights viruses.
- B cells target complex 3D structures, while T cells recognize simpler protein fragments.
Sources
- Cargill, Michael. “Antigen Encounter and Presentation,” “B Cell Development and Function,” “T Cell Development and Function.” Immunology, North Carolina School of Science and Mathematics, Durham, North Carolina, April 2024.
- DeRoo, J., Terry, J. S., Zhao, N., Stasevich, T. J., Snow, C. D., & Geiss, B. J. (2024). Pabfold: Linear antibody epitope prediction using AlphaFold2. eLife. https://doi.org/10.7554/elife.98369
- Peters, Bjoern. “B Cell Epitope Prediction.” B Cell Epitope Prediction, 2020 User Workshop – 2.6, Immune Epitope Database and Analysis Resource, Virtual, November 5-6, 2020.
- Ranganathan, S. (2019). Encyclopedia of Bioinformatics and Computational Biology. volume 1, methods. Elsevier
- Ras-Carmona, A., Lehmann, A. A., Lehmann, P. V., & Reche, P. A. (2022). “Prediction of B cell epitopes in proteins using a novel sequence similarity-based method.” Scientific Reports, 12(1). https://doi.org/10.1038/s41598-022-18021-1
- Sanchez-Trincado, J. L., Gomez-Perosanz, M., & Reche, P. A. (2017). “Fundamentals and methods for T- and B-cell epitope prediction.” Journal of Immunology Research, 2017, 1–14. https://doi.org/10.1155/2017/2680160
- Schaap-Johansen, A.-L., Vujović, M., Borch, A., Hadrup, S. R., & Marcatili, P. (2021). “T cell epitope prediction and its application to immunotherapy.” Frontiers in Immunology, 12. https://doi.org/10.3389/fimmu.2021.712488
- Sette, Alessandro. Interview. Conducted by Teresa Fang, 22 Jul. 2024
Editorial Team
- Chief Editor: Katherine Mi
- Associate Editor: Christine Chen
- Team Editor: Chloe Eng
- Image Credit: Teresa Fang
- Social Media Lead: Chloe Eng
Mentor
- Rasika Vartak, Ph.D. is a molecular biologist and neuroscientist. She is currently working at the Quantitative Biosciences Institute at the University of California, San Francisco.
Content Expert
Dr. Alessandro Sette is a professor at the La Jolla Institute for Immunology (LJI) in San Diego, California, and the Director of the Center for Cancer Immunotherapy and Center for Vaccine Innovation. He is part of a team that develops and oversees the national Immune Epitope Database (IEDB), a freely available, widely-used bioinformatics resource database for storing epitope structures. In 2020, Dr. Sette published the first study on SARS-CoV-2 epitopes that are targeted by the human immune system and has co-led research into T-cell responses in COVID-19 patients and cross-reactivity to other coronavirus epitopes.